Yes I’m saving 2500 points to check if the orbits is fine. Sometimes I just need the last point than I save the last point.
I tried to use StaticArrays however i’m not sure what i was doing wrong or if it’s even possible use in systems like this.
In that case i’m solving the CR3BP equations with the variational equations and a chaotic indicator
For 5e3 periods of my orbit i’m getting 2.47GB memory usage avg
For 5e4 periods its 24.6GB
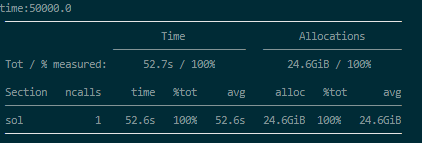
Perhabs its already maximum optimized but i’m not sure. i’ll share my code down there.
using Plots, LinearAlgebra, Random, DataFrames, TimerOutputs, StaticArrays, DifferentialEquations
const to = TimerOutput()
@inline @inbounds function vy0(C,p,v)
"""
Calculate Vy0 from jacobian value
input : (C,p,v) ...
C = Jacobi cte
v = x,y,vx
p = mass ratio parameters mu
"""
#cartesian coordinates
x, y, x̂= v
#distance vectors
r1 = ((x+μ)^2 + y^2)^(1/2)
r2 = ((1-x-μ)^2 + y^2)^(1/2)
#jacobi integral - constant C.
ŷ = (x^2 + y^2 - x̂^2 + 2*((1-μ)/r1 + μ/r2) - C)^(1/2)
return ŷ
end
"""
Initial Conditions
"""
global μ = 0.01 # mass ratio binary main system
# third body
CJ = 0 # Jacobi cte
x0 = 1.893
y0 = 0
vx0 = 0
vyu = -vy0(CJ,μ,[x0,y0,vx0]) #sign negative for retrograde orbits
"""
Create a random set of tangent vector for MEGNO
Get normalized vectors for MEGNO
"""
rng = MersenneTwister(234);
a1 = rand!(rng, zeros(4))
global dd = 0
for i in 1:length(a1)
if a1[i] <= 0.5
a1[i] = -a1[i]
end
end
for i in 1:length(a1)
global dd += a1[i]^2
end
dd = sqrt(dd)
for i in 1:length(a1)
a1[i]= a1[i]/dd
global dd += a1[i]^2
end
####################
"""
u0 state vector = [x0,y0,vx0,vy0,xrng,yrng,vxrng,vyrng,megno_mean,megno_final]
x0,y0,vx0,vy0 -> Third body
%%%% MEGNO - REFERENCE : https://www.aanda.org/articles/aa/pdf/2001/41/aah2931.pdf
xrng,yrng,vxrng,vyrng = random initial conditions to calculate megno with variational equations
megno_mean = mean value of (dot var_equations / var_equations)
megno_final = numerical integration of the MEGNO mean value
"""
u0 = [x0, y0, vx0, vyu, a1[1], a1[2], a1[3], a1[4], 0.0, 0.0] # vector with initial conditions
println("Initial state vector :",u0)
println("JacobiCte Initial :",CJ)
# Time Span
t_end = 2*pi*5e4
tspan=(0.0,t_end)
numberofpoints = 2500
@inline @inbounds function cr3bp_rhs(du,u,μ,t)
"""
Planar Circular Restricted 3body problem
Equations of motion in co-rotating frame
in:
u = position and velocities of the particle
μ = mass ratio parameter == μ2
t = time span
out:
du = integration of the acel. and vel.
"""
#cartesian coordinates
r1 = ((u[1] + μ)^2 + u[2]^2)^(1/2)
r2 = ((-1 + u[1] + μ)^2 + u[2]^2)^(1/2)
du[1] = u[3] # F1
du[2] = u[4] # F2
du[3] = 2*u[4] + u[1] - (1 - μ)*(μ + u[1])/(r1^3) - μ*(-1 + μ + u[1])/(r2^3) # F3
du[4] = -2*u[3] + u[2] - (1 - μ)*u[2]/(r1^3) - μ*u[2]/(r2^3) # F4
# renormalizing tangent vectors everystep for MEGNO - Chaotic Indicator
normvec = sqrt(u[5]^2 + u[6]^2 + u[7]^2 + u[8]^2)
xm = u[5]/normvec
ym = u[6]/normvec
vym = u[7]/normvec
vxm = u[8]/normvec
# mass parameters
mu1 = 1 - μ
mu2 = μ
#derivatives f3
df3x = 1.0-mu1/r1^3-mu2/r2^3+3.0*mu1*(u[1]+mu2)^2/r1^5+3.0*mu2*(u[1]-mu1)^2/r2^5
df3y = 3.0*mu1*(u[1]+mu2)*u[2]/r1^5+3.0*mu2*(u[1]-mu1)*u[2]/r2^5
df3vx = 0
df3vy = 2
#derivatives f4
df4x = 3.0*mu1*(u[1]+mu2)*u[2]/r1^5+3.0*mu2*(u[1]-mu1)*u[2]/r2^5
df4y = 1.0-mu1/r1^3-mu2/r2^3+3.0*mu1*(u[2])^2/r1^5+3.0*mu2*(u[2])^2/r2^5
df4vx = -2
df4vy = 0
du[5] = vxm
du[6] = vym
du[7] = df3x*xm + df3y*ym + df3vx*vxm + df3vy*vym
du[8] = df4x*xm + df4y*ym + df4vx*vxm + df4vy*vym
#megno evolve
f = u[5]*du[5] + u[6]*du[6] + u[7]*du[7] + u[8]*du[8]
d2 = u[5]^2 + u[6]^2 + u[7]^2 + u[8]^2
# megno equations
if t==0 # need check at t=0 to avoid overflows caused by 0 on the denominator
chaosind1 = (f/d2) * (t + 1e-16)
du[9] = chaosind1
du[10] = 2*u[9]/(t + 1e-16)
else
chaosind1 = (f/d2) * t
du[9] = chaosind1
du[10] = 2*u[9]/t
end
end
# solver
p = μ
prob = ODEProblem(cr3bp_rhs,u0,tspan,p)
@timeit to "sol" solution2 = solve(prob,Vern9(),reltol = 1e-16,abstol = 1e-16, maxiters = 1e8, saveat = t_end/numberofpoints)
df = DataFrame(solution2)
println("points:",length(df.timestamp))
println("time:",df.timestamp[end]/(2*pi))
"""
Checking if the cte error increases through the time
"""
jacobicte = []
for i in 1:length(df.timestamp)
x= df.value1[i]
y= df.value2[i]
vx= df.value3[i]
vy= df.value4[i]
r1 = ((x + μ)^2 + y^2)^(1/2)
r2 = ((-1 + x + μ)^2 + y^2)^(1/2)
jacobi = (x^2 + y^2 - vx^2 - vy^2 + 2*((1-μ)/r1 + μ/r2))
append!(jacobicte,jacobi)
end
p1 = scatter(df.timestamp,jacobicte, label="Jacobi Constant")
p2 = scatter(df.value1,df.value2)
print_timer(to::TimerOutput)
#p4 = plot(df.value9)
np= []
np2 = []
for i in 1:length(df.timestamp)
b= df.value10[i]/df.timestamp[i]
append!(np,b)
end
p3 = plot(df.timestamp,np, label=" MEGNO <Y>", legend=:bottomright)
println("FINAL MEGNO: ", np[end])
plot(p1,p2,p3)

